Tutorial: The R Programming Language
by Kevin Kotzé
The original creation of R was undertaken by Gentleman & Ihaka at the University of Auckland during the early 1990’s. The continuing development of this open source programming language has since been taken over by an international team of academics, computer programmers, statisticians and mathematicians. One of the key features of this environment is that it allows the user to program algorithms and use tools that have been programmed by other users, after they have created a dedicated language. For example, there are several thousand packages on the Comprehensive R Archive Network (CRAN) and there are many more on other repositories, such as GitLab, GitHub, BitBucket or personal webpages. In addition, the full source code for R has been made avaiable to all users who can inspect, modify and extend it to their needs.
Although much of the development of the base code is performed on Linux systems, the software can be run on any platform and it allows users to write functions, do calculations, apply most statistical and mathematical techniques, create simple or complicated graphs, or even write a personalised library of functions. It is able to work with extremely large datasets and also handles a wide variety of data types including scalars, vectors, matrices, data frames, lists, objects, etc. Furthermore, there are a number of interfaces that have been developed which allow for R to work with other programs, source code, or databases (including Excel, Matlab, Stata, Python, Octave, Bloomberg, Datastream, C++, Fortran, SQL, Oracle, etc.).
While R was originally developed as a statistics software package, it has evolved into much more. You can now use it as a pure mathematical tool as there are several packages that facilitate analytical derivations with the aid of symbolic and numeric computation (in addition to a number of other techniques). This allows for one to solve complex nonlinear systems of equations, derivatives, integrals, etc. In addition, one could also use it to organise, or rearrange large datasets, and as a result it is often used on large commercial “big data” or “data science” projects.
Many research institutes, companies, and universities have migrated to R and most modern textbooks now containing references to R source code, where all of the calculations make use of R functions. In 2013 Oracle estimated that there were over 2 million users (although this number has grown exponentially since then) and in a recent article in the New York Times, Google economist Hal Varian noted that there has been a significant growth in popularity of the R language as a tool for data analysis.
Furthermore, since it is both free and extremely powerful, most employers are loath to spend money on commercial software, such as Matlab or Stata; so there is a good chance that you will no longer be able to work with these software platforms when you enter the workforce.
1 Installing R on your machine
To download and install R for the first time you can go the webpage:
Select the platform for your machine (i.e. Windows) and select: install R for the first time. Thereafter, select the latest version of the software and install as you would for any other program. If you are making use of a linux operating system then go to https://cran.r-project.org/bin/linux/, select your distribution (i.e. debian, redhat, suse, or ubuntu) and follow the instructions that are provided.
While the base R software comes with a GUI, it is very basic and not ideal for someone who is starting out. As such we are going to make use of the RStudio integrated development environment (IDE), which can be downloaded for free at:
To select the download the user interface go to the RStudio website and select the appropriate options to download the desktop version of this software. After you have installed this software, you will then be able to gain access to the wonderful world of R through the RStudio application.
While RStudio is possibly the most popular IDE for R, you may wish to make use of generic IDE’s such as Virtual Studio, Atom, Sublime Text, Kate, Notepad ++, or Vim. These are all pretty useful if you are simultaneously coding in different software languages, or if you want to make use of a single environment to do your modelling and description of the model (although there are now better ways of doing this within the R programming environment).
In addition to these two programs, I would also recommend that Windows users should download and install the Rtools program, which is available at:
https://cran.r-project.org/bin/windows/Rtools/
Apple mac users should consider installing the Xcode command line tools, which are available at:
https://developer.apple.com/downloads
Linux users should install the R development package, usually called r-devel or r-base-dev.
This will allow you to interface with GitLab or GitHub accounts and would allow you to gain access to many other interesting aspects of this programming language.
2 Resources
You can find helpful tutorials by following CRAN’s link to Contributed Documentation. If you are new to this language, then Paradis (2005) is a great introduction. The English version can be download from the CRAN website:
http://cran.r-project.org/doc/contrib/Paradis-rdebuts_en.pdf
More recently, there are also a number of online courses that provide a great introduction. For example, you may wish to take a look at:
https://www.datacamp.com/courses/free-introduction-to-r
Or alternatively, all UCT staff and students can access the online training material at https://www.linkedin.com/learning at no charge, where you may want to consider taking the courses:
https://www.linkedin.com/learning/learning-r/welcome?u=70295562
https://www.linkedin.com/learning/r-statistics-essential-training/welcome?u=70295562
https://www.linkedin.com/learning/data-science-foundations-fundamentals/exercise-files?u=70295562
Other options include teaching yourself R with the aid of the swirl package that has been written in R. Details of this package can be found at:
For those who would like to invest more time on programming, then you could take a look at the text by Wickham (2014). In addition, his website is particularly good and his GitHub repository includes a few GitBook’s that are worth reading. One of which is the text:
He is also the author of the tidyverse, which is the main package for data wrnagling and visualisation. You can check out details of this at: https://www.tidyverse.org/.
The academic publisher Springer has released an inexpensive and focused series of books, titled Use R!, which discuss the use of R in a various subject areas. The series now includes over 65 titles and they all contain source code, which I have found these to be extremely useful. You can find these titles at:
http://www.springer.com/series/6991?detailsPage=titles
The publisher CRC Press (which is part of the Taylor & Francis Group) have a similar series of books, which may be viewed at:
https://www.crcpress.com/Chapman--HallCRC-The-R-Series/book-series/CRCTHERSER
A further useful resource is the Journal of Statistical Software, which contains journal articles that seek to explain many of the packages that are available on the R website. These articles are usually similar to the vignettes that accompany most packages.
For a list of the major econometric type of packages, including those that have been developed for time series, microeconometric and panel models, see the websites:
http://cran.r-project.org/web/views/Econometrics.html
https://cran.r-project.org/web/views/TimeSeries.html
https://cran.r-project.org/web/views/Finance.html
If you are also interested in computational issues, then you may wish to visit the webpages:
https://cran.r-project.org/web/views/NumericalMathematics.html
https://cran.r-project.org/web/views/HighPerformanceComputing.html
Alternatively for the complete set of the package categories:
https://cran.r-project.org/web/views/
Or you can find all the packages that have been sorted by name at:
http://cran.r-project.org/web/packages/available_packages_by_name.html
Remember also that the total number of packages on the CRAN repository represent only a small fraction of those that are available. So if you don’t find what you like on this particular repository then it may be available elsewhere, possibly on a GitHub or GitLab repository, you just need to search.
In addition, if you are struggling with a particular function in R, then you may want to check out the StackOverFlow webpage, which was originally developed for computer programmers (many of which make use of R for data science applications). The probability that your query has been previously answered is usually quite high and what is particularly nice about this website is that it has a number of methods that avoid the duplication of queries. You can check it out at:
Then lastly, as most of my programming experience has been obtained in Matlab, I have also found the text by Hiebeler (2015) to be of use. He has a particularly nice summary document that can be found at:
http://www.math.umaine.edu/~hiebeler/comp/matlabR.pdf
Similar documentation can be found for those who may be more familiar with Python or Julia.1 Unfortunately, I have not found anything that is all that good for Stata users, as the Stata environment is somewhat different to that of most other scripting languages. However, I can tell you that some of the Stata users that I’ve worked with have found the transition to R to be relatively straightfoward. Those who are interested may find the book by Muenchen and Hilbe (2010) to be of use, although it is not a quick read.2 If you are interested in using Python for econometrics then check out the website of Kevin Shepphard at:
In addition, if you want to use either Python or Julia for applications in mathematical economics then check out the website of Thomas Sargent and John Stachurski at:
3 Desktop Overview
After you have installed the software that has been mentioned above, you will be able to open RStudio. When making use of this software, it would look similar to that which is shown in Figure 1.
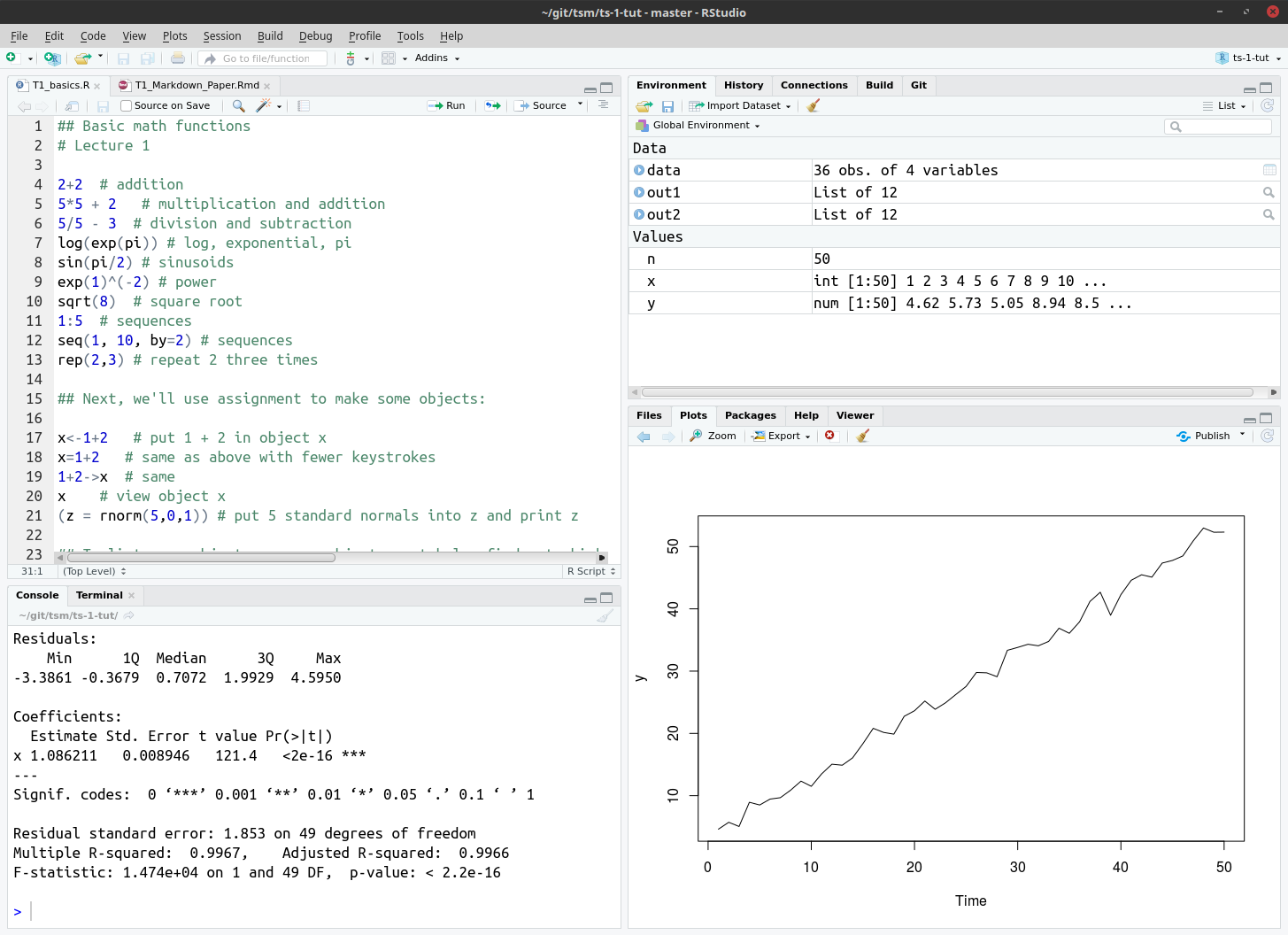
- The basic program file (what Stata users call do files) is usually displayed in the top left-hand corner. One is able to view multiple program files at once and you can also execute entire files, single lines or just a subset of a line.
- All the variables, matrices, scalars, objects, strings, etc. are store in the
Environmenton the top right. - Any conceivable type of graph may be constructed and displayed in the
Plotspanel on the bottom right. These may be exported as jpeg’s, pdf’s, etc. - The
Consoleon the bottom left may be used to input commands or provide output (which may be structured to your liking). - If you double-click on a Data element in the
Environmentit will display it in a Excel type screen on the left panel. - The additional tabs on the bottom right contain additional useful information. Screen shots of these are contained below.
- The
Filesdirectory takes on a Windows Explorer type of interface, which allows you to negotiate your way around your current directory. - There is a very useful
Helpfunction that provides the exact format for the respective commands. It will also contain an example of it’s use as well as textbook/journal references (where appropriate). - To install and use
Packagesyou can use the respective tab, which will link you to the repository on the web. When you click on the package name you will see all the available functions in the package. In addition, some packages will include a very useful vignette, which is an equivalent of a journal article that explains the methodology and application of the package. It will also include a working example with code (in most cases).
4 Basic overview of programming in R
#> Basic commands: ----
2 + 2 # addition## [1] 45*5 + 2 # multiplication and addition## [1] 275/5 - 3 # division and subtraction## [1] -2log(exp(pi)) # log, exponential, pi## [1] 3.141593sin(pi/2) # sinusoids## [1] 1exp(1)^(-2) # power## [1] 0.1353353sqrt(8) # square root## [1] 2.8284271:5 # sequences## [1] 1 2 3 4 5seq(1, 10, by=2) # sequences## [1] 1 3 5 7 9rep(2,3) # repeat 2 three times## [1] 2 2 2#> Assign values to objects: ----
x <- 1 + 2 # put 1 + 2 in object x
x = 1 + 2 # same as above with fewer keystrokes
x # view object x## [1] 3(z = rnorm(5,0,1)) # put 5 standard normals into z and print## [1] 0.7241835 -1.1344268 0.7276477 -1.3847352
## [5] -1.6150733#> List objects, get help and get current working directory: ----
ls() # list all objects## [1] "figs" "tbls" "x" "z"help(exp) # specific help (?exp is the same)
getwd() # get working directory (tells you where R is pointing)## [1] "/home/kevin/git/tsm/ts-1-tut"#> Set working directory and quit: ----
setwd('C://Users') # change working directory (note double slash)
q() # end the session (keep reading)#> To create your own data set: ----
mydata = c(1,2,3,2,1) # where we concatenated the numbers
mydata # display the data## [1] 1 2 3 2 1mydata[3] # the third element## [1] 3mydata[3:5] # elements three through five## [1] 3 2 1mydata[-(1:2)] # everything except the first two elements## [1] 3 2 1length(mydata) # length of elements## [1] 5mydata = as.matrix(mydata) # make it a matrix
dim(mydata) # provide dimensions of matrix## [1] 5 1#> Rules for doing arithmetic with vectors: ----
x=c(1,2,3,4); y=c(2,4,6,8); z=c(10,20) # three commands are stacked with ;
x*y # i.e. 1*2, 2*4, 3*6, 4*8## [1] 2 8 18 32x/z # i.e. 1/10, 2/20, 3/10, 4/20## [1] 0.1 0.1 0.3 0.2x+z # i.e. 1+10, 2+20, 3+10, 4+20## [1] 11 22 13 24x %*% y # matrix multiplication## [,1]
## [1,] 60#> Basic loops: ----
for (id1 in 1:4){
print(paste("Execute model number", id1))
} # for loops are the most common## [1] "Execute model number 1"
## [1] "Execute model number 2"
## [1] "Execute model number 3"
## [1] "Execute model number 4"id2 <- 1
while (id2 < 6) {
print(id2)
id2 = id2 + 1
} # while loops can be used as an alternative## [1] 1
## [1] 2
## [1] 3
## [1] 4
## [1] 5x <- -5 # use else statement to test if this is positive or negative
if (x == 0) {
print("Zero")
} else if (x > 0) {
print("Positive number")
} else {
print("Negative number")
}## [1] "Negative number"# Could also vectorise code and use `sapply or `tidyverse::map` function to make code run quicker#> Simulating regression data: ----
n <- 50 # number of observations
x <- seq(1, n) # right-hand side variable
y <- 3 + x + rnorm(n) # left-hand side variable
plot(x, y) # show the linear relationship between the variables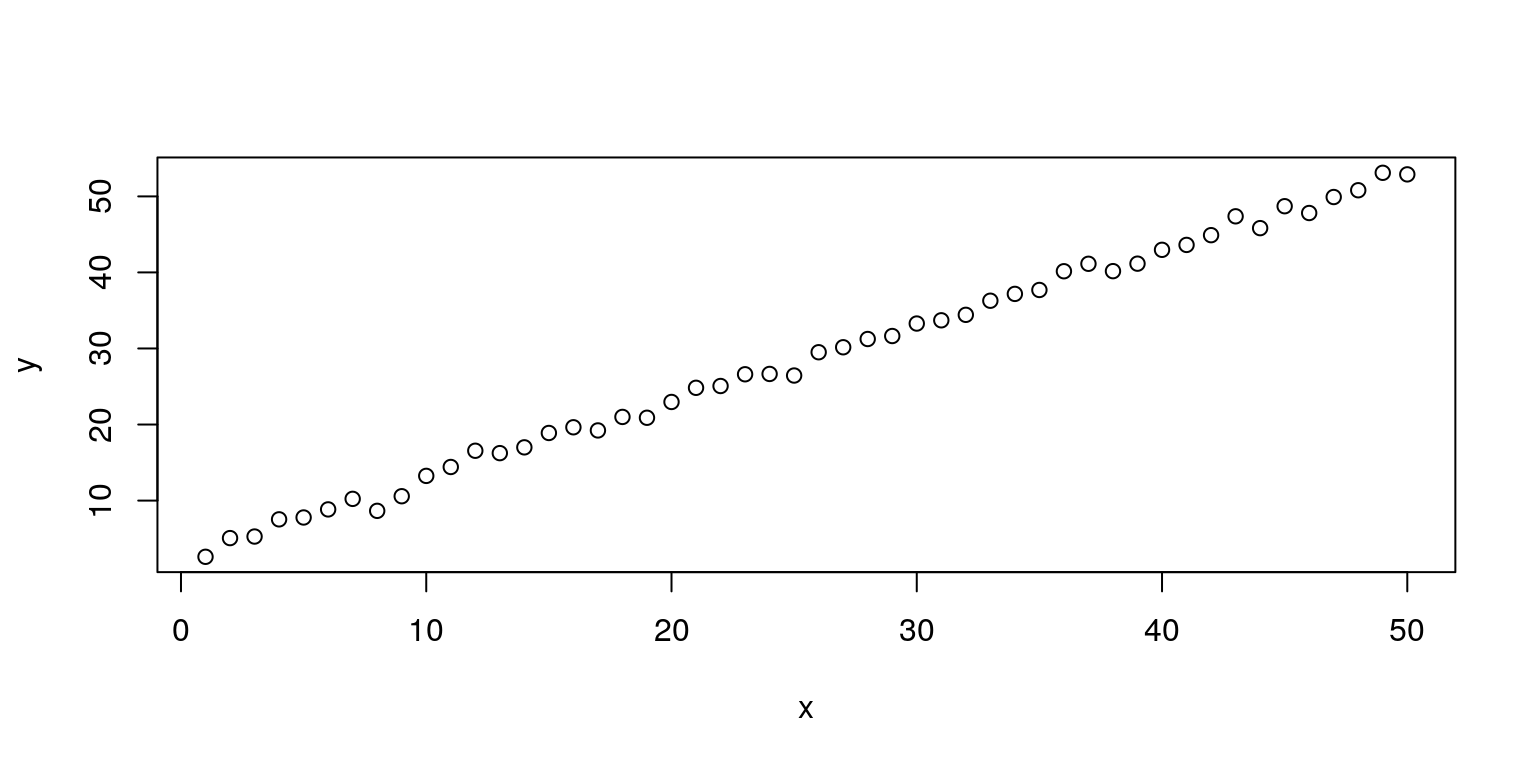
plot.ts(y) # plot time series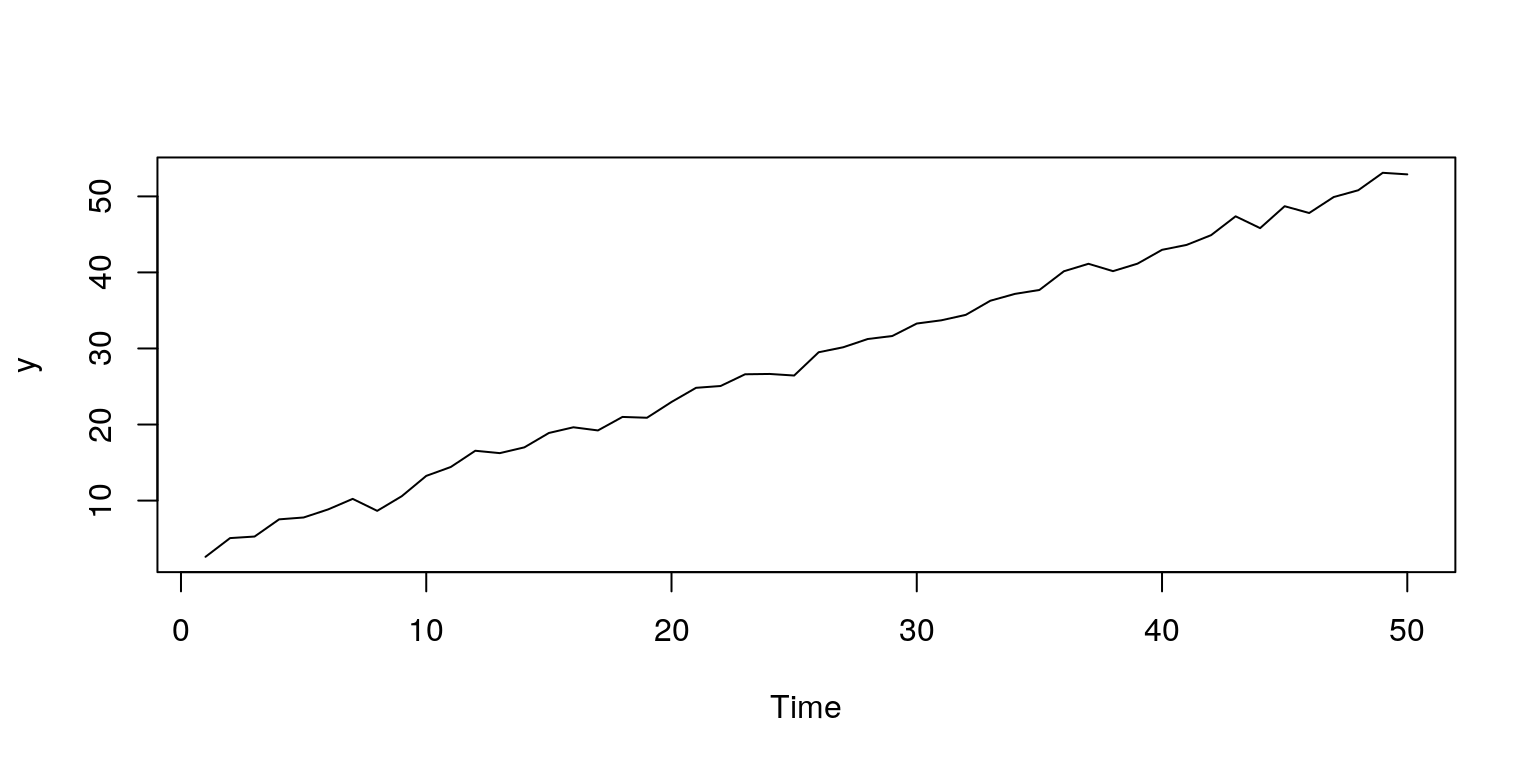
#> Estimate a linear regression model: ----
out1 <- lm(y ~ x- 1) # regress x on y without constant
summary(out1) # generate output##
## Call:
## lm(formula = y ~ x - 1)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.5994 -0.3817 1.1129 1.7931 5.0407
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## x 1.087158 0.009542 113.9 <2e-16 ***
## ---
## Signif. codes:
## 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.977 on 49 degrees of freedom
## Multiple R-squared: 0.9962, Adjusted R-squared: 0.9962
## F-statistic: 1.298e+04 on 1 and 49 DF, p-value: < 2.2e-16#> Working with real data: ----
rm(list=ls()) # clear data
graphics.off() # close graphssetwd('C:\\TimeSeries\\1_Intro\\tut')data <- read.csv(file="data.csv") # read in csv dataout2 <- lm(data$y ~ data$k) # estimation a linear regression
summary(out2) # generate output##
## Call:
## lm(formula = data$y ~ data$k)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.51371 -0.23632 0.06197 0.25068 0.40872
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.63668 0.09998 6.368 2.88e-07 ***
## data$k -0.14167 0.17601 -0.805 0.426
## ---
## Signif. codes:
## 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.3093 on 34 degrees of freedom
## Multiple R-squared: 0.0187, Adjusted R-squared: -0.01016
## F-statistic: 0.6479 on 1 and 34 DF, p-value: 0.4265# clear data & close graphs
rm(list=ls())
graphics.off()5 Loading data
The easiest way to import data into R is to make use of a .csv file format from an Excel spredsheet. You can take a look at the existing datafiles that I use in the tutorials to get an idea of the suggested format. Note that I use columns for the data and would give the column a name in the first cell of the column.
Once you have your data as you want it in Excel you need to then save it. Choose the option to File > Save As, then under the type of file, change the Excel Workbook (.xlsx) to CSV (Comma delimited)(.csv). In this example, I have called my data file data.csv.
Now you can go to RStudio, where you will see in each program file I would always clear the data and close the plots with the following commands.
rm(list=ls()) # clear data
graphics.off() # close graphsThereafter, it is a good idea to specify a working directory, which would be where you data file is located.
setwd('C:\\TimeSeriesAnalysis\\1_Intro\\tut')The next part would then be to import the data with the use of read.csv command. In this case, we create an object called data that will have the column names for each of the time series variables.
data <- read.csv(file="data.csv") # read in csv dataIn some versions of Windows (notably Windows 8) it saves a .csv file with semi-comma separators, in which case you would need to change the above command to:
data <- read.csv(file="data.csv", sep=";") # read in csv dataRemember also that you need to make sure that the variable has been correctly loaded by plotting the data. In some instances the data may be imported as a logical operator or as a factor. To plot the data make use of the command:
plot.ts(data$y) # plot time serieswhere the variable in our Excel datafile would have been labelled as y. If it looks incorrect then you can always convert the variable into a numeric field using the command:
data.tmp <- as.numeric(data$y)In this case the new variable would be called data.tmp. As an alternative to using these commands, you can just use the Import Dataset button on the Environment tab and it will take you through the necessary steps.
6 Other aspects of interest
6.1 Reproducible research
One of the major advantages of R is that people have written programs that allow for it to interface with other languages. This could be used to generate a reproducible research, where you can write your entire paper, book or slides within the RStudio environment. Two of the important packages that allow you to do this are rmarkdown and knitr, where the knitr package may be used to knit the RMarkdown documents to create HTML, PDF, MS Word, LaTeX Beamer presentations, HTML5 slides, Tufte-style handouts, books, dashboards, shiny applications, scientific articles, websites, and more.
For more on the use of rmarkdown and knitr, see Xie (2013) and the website http://rmarkdown.rstudio.com/.
The idea behind these forms of reproducible research programs is that the text for your report (or article) would be written in standard LaTeX and the model would be included as a R chunk of code. This file could then be compiled within R to generate a .md (which is useful for web documents), .pdf or .tex file. The knitr package has also been integrated with the LaTeX editor LyX, which allows you to produce reproducible research files from within this environment. More complex documents, such as books, could be written with the bookdown package. An example of a basic Rmarkdown document is included under separate file.
6.2 Speeding up R
While R does not perform computations at the speed of other lower level coding environments (such as C++, C, or Fortran) it does have the ability to interface with these (and other) languages. In particular one could make use of the Rcpp package, for which details are provided in Eddelbuettel (2013). In addition, there are also numerous parallel processing packages that could be used, the most popular of which is probably snow.
One could also make use of the graphics processor unit (GPU) in your machine to assist with the computations, by incorporating Cuda functions if you have a Nvidia GPU. Then lastly, as this is freeware, you could also make use of a cloud server or a cluster of cloud servers to perform massive computations in R with relative ease as it would not be necessary to purchase any software for each instance that you would need to make use of. A number of companies have already established such servers that are preloaded with R for your ease of use, where you only pay for the computational time that you make use of. For example, see:
Of course, you could also set this up yourself with the aid of several servers on Amazon Web Service (AWS), which would allow you access to an impressive amount of computing power at a relatively low cost (i.e. there is even a free tier option). This is actually relatively easy to do and the computing power that you are able to use is impressive, although you need to be aware of the costs of using large machines. You can find a brief tutorial on how to make use of these cloud computing resources (in what I think is the easiest possible way) on my website at:
6.3 Developing with GitLab, BitBucket or GitHub
GitLab, BitBucket and GitHub are web-based repositories that allow for the source code management (SCM) functionality of Git as well as many other features. It can be used to track all the changes to programming files by any oermitted user, so that you can see who may have made a change to a particular file. In addition, if it turns out that a particular branch of code did not provide the intended result, then you can always go back to a previous version of the code. Most R developers make use of either GitLab or GitHub for the development of their packages and it is particularly useful when you have many developers working on a single project.
For your own personal work you should consider opening a GitLab account, which will allow you to store public and private repositories for free. As an alternative, you can make use of GitHub for a free public repository, or BitBucket, which provides users with free private repositories. If you want to get more involved in programming then you are able to download all of the packages that you may want to load the GitKraken or GitHub Desktop software onto your machine, as it will assist with most of the uploading and downloading of files, maintenance, etc.
After installing the Rtools software one is also able to download and install packages that are located on GitLab, BitBucket or GitHub accounts, directly from within R with the aid of the devtools package. For example, the following code would find the package on GitHub, provided you are connected to the internet:
library("devtools")
install_github("username/packagename")Since we only usually make use of this single command from within the devtools package we could also write this command as:
devtools::install_github("username/packagename")Or alternatively, if you have GitHub Desktop then you could clone the repository and make use of the following code to install the package from you hard-drive:
install.packages("C:\\GitHub\\ggplot2", repos=NULL, type="source")Then lastly, if you are developing an R package then you may wish to make use to create a R-project in RStudio so that you can control the uploads and downloads to the GitHub repository from within RStudio.
For a brief tutorial on the GitHub system, you can take a look at the following document that is located on my website:
6.4 Updating R and the associated packages
On average the based code and all of the packages in R are updated about three times a year. Now if you have installed a large number of packages, this could be quite a time consuming exercise. The easiest way that I have found to perform this task is to firstly install the new version of R and RStudio; where lately I’ve been using the multithreaded BLAS/LAPACK libraries that are included in the Microsoft R Open (MRO) distribution that may be accessed through https://mran.microsoft.com/open.
If it is a minor release, such as a change from 3.4.0 to 3.4.1 then you can just open the Packages tab on RStudio and click on update packages and everything will be taken care of. However, when they provide a major release, such as a change from 3.3.5 to 3.4.0, then you need to do a bit more work. The easiest way that I’ve found to update all the packages is to firstly find out where all your packages are stored with the following command:
.libPaths()Thereafter, I would use Windows Explorer (or Finder if using a Mac) to copy the folder 3.3.5 and paste it in the same parent directory. The copy of the folder would then need to be renamed 3.4. The last step is to go back to RStudio and run the command:
update.packages()And it will do everything for you.
7 Concluding Comments
During the course of this semester we are really only going to touch the surface of what is possible in this programming environment. For those of you that want to explore the data-organising capabilities, then start by taking a look at the tidyverse package for data wrangling and if you need to interact with a large database, the RODBC package is simple to use and works nicely. If you are working with time series data that has strange frequencies then the zoo or xts packages are particularly useful. For working with time series data the lubridate and tibbletime packages allow us to manipulate large datasets with relative ease.
In addition, if you want to make attractive figures for your research then the ggplot2 package should be a good first port of call. And if you want to make use of an interactive website then the shiny package has impressive functionality. And then there are any number of statistical and machine learning packages that could be used for most forms of analysis.
8 References
Eddelbuettel, Dirk. 2013. Seamless R and C++ Integration with Rcpp. UseR! Series. Springer.
Hiebeler, David E. 2015. R and Matlab. New York: Chapman & Hall/CRC.
Muenchen, Robert A., and Joseph M. Hilbe. 2010. R for Stata Users. New York: Springer.
Paradis, Emmanuel. 2005. “R for Beginners.” Mimeo. Université Montpellier II.
Wickham, Hadley. 2014. Advanced R. New York: Chapman; Hall/CRC.
Xie, Yihui. 2013. Dynamic Documents with R and Knitr. New York: Chapman; Hall/CRC.
These open source languages are also developing at a rapid rate and are excellent alternatives to R. What is particularly nice about Python is that it includes relatively extensive symbolic and numerical computational packages (but possibly not as many statistical functions). Julia allows for more expedient computation, but requires a slightly higher level of programming skill (i.e. somewhere between C++ and Python).↩
This is available at the UCT library.↩